Patrick McDeed is awarded a Fred Hutch Mahan Fellowship. Patrick joined the Ha Lab in June 2025 as a postdoc. His project is titled “ctDNA-based modeling of genotypic & phenotypic dynamics during mCRPC therapy”. He aims to develop new computational methods and statistical frameworks for the analysis of circulating tumor DNA (ctDNA) obtained from cancer patients as non-invasive “liquid biopsy” to identify genotypic and phenotypic features that can better monitor treatment response and predict therapeutic outcomes. The development of these ctDNA-based prediction and serial monitoring tools will provide insights to improve treatment strategies and management of prostate cancer patients receiving Lutetium-PSMA radioligand therapies.

Samantha Schuster is awarded a Fred Hutch Translational Data Science Postdoc Fellowship. This is two-year award, which will provide funding support for her salary and research. Samantha joined the Ha Lab in February 2024 as a postdoc jointly mentored by Gavin Ha and Dr. Andrew Hsieh. Her research is focused on studying the tumor heterogeneity and evolution in advanced bladder cancer from whole genome sequencing, circulating tumor DNA, and single-cell RNAseq analyses. She is also investigating the mechansims of resistance to therapies, such cisplatin chemotherapy, and their differences between histological subtypes. The outcome of this work will characterize the advanced bladder cancer to improve our understanding of why rare subtypes are more aggressive with poorer outcomes.

Congratulations to Robert Patton on receiving a 2023 Prostate Cancer Foundation Young Investigator Award.
Robert’s project titled “Non-Invasive Prediction of Tumor Phenotypes, Therapeutic Targets, and Treatment Response in CRPC using ctDNA” proposes to develop novel computational approaches to study of prostate cancer phenotypes and treatment response using liquid biopsies. The outcome of his work will have important applications in tumor monitoring, predictive biomarker discovery, and diagnostics for men with advanced prostate cancer.

Thomas Persse joins the Ha Lab as a Bioinformatics Analyst. He holds a Master’s in Bioinformatics and Genomics and has industry experience working on cell-free DNA sequencing analysis.
Armand Bankhead is a joint Senior Staff Scientist with the Nelson Lab. He holds a PhD in Bioinformatics and Computational Biology with over 15 years of experience in academia and industry working on many topics in cancer research, including cancer genomics and circulating tumor DNA analysis.

Manasvita Vashisth joins as a postdoctoral fellow. She completed her PhD at the University of Pennsylvania in Chemical and Biomolecular Engineering. She joins the Ha Lab at Fred Hutch to study genomic and phenotypic heterogeneity in prostate cancer from tissue and circulating tumor DNA.

We are excited to welcome Addison Gage and Akira Nair who join the lab as summer research interns in computational biology.
Addison will work on implementing the lab’s tools into the cloud environment and exploring maching learning approaches for cancer genomics.
Akira is part of the Fred Hutch Summer Undegraduate Research Program (SURP). He will be applying genomic analyses and deep learning approaches to analyze circulating tumor DNA.

Welcome to Sarah Huang who joins the lab as a rotation student from the MCB Graduate Program. Sarah will be developing deep learning approaches to study gene transcription activity in circulating tumor DNA.

Welcome to Katherine Feldmann who joins the lab as a rotation student from the MCB Graduate Program. Katherine will be investigating cancer genome structure using the new T2T reference genome and long-read sequencing.

Welcome to Michael Yang, who joins the lab as a new Bioinformatics Analyst. He recently completed a Master’s in Biomedical Engineering at Columbia University, specializing in Bioinformatics and Data Science.

We are very grateful to the NIH for supporting the lab with the 2022 NIH Director’s New Innovator Award. The DP2 New Innovator Award will support the Ha Lab for the next 5 years to explore high-risk, high-reward research into how multiomic (genomic, transcriptomic, and epigenetic) information can be exploited from liquid biopsies to improve clinical applications and precision oncology.

Anna-Lisa Doebley successfully defended her PhD dissertation titled “Predicting cancer subtypes from nucleosome profiling of cell-free DNA”. Anna-Lisa capped off 3.5 years of research in liquid biopsies and circulating tumor DNA with an amazing defense presentation. Special thanks for co-mentor David MacPherson and supervisory committee members Pete Nelson, Stephen Tapscott, and Trevor Bedford

Pushpa Itagi is awarded a Fred Hutch Translational Data Science Postdoc Fellowship, which will provide funding support for her salary and research. Pushpa proposed to work on elucidating metastatic bladder cancer (mBLCA) evolution from rapid autopsy tissue samples. She will use statistical and machine learning methods to decipher cancer spread from the bladder to distant tissues. Using a combination of other data types such as methylation, expression, and cell-free DNA from the patients she intends to provide an extensive multi-comics characterization for mBLCA tumor heterogeneity. Pushpa is jointly mentored by Dr. Andrew Hsieh. She interface with other key collaborators in the Bladder Cancer Groups at Fred Hutch and UW.

Welcome to rotation students, David Chen and Bhargav Vemuri.
David joins the lab as graduate rotation student in the WWAMI program. He is a 2nd year medical student in the UW School of Medicine. He will be working on a project to study response to radiation therapy from prostate cancer patients using cell-free DNA.
Bhargav is a first year student in the UW Biomedical and Health Informatics graduate program and joins us for his Summer 2022 Rotation. Bhargav will be working on studying genomic heterogeneity in bladder cancer.

The labs of Gavin Ha and David MacPherson receive a 3-year grant from the Kuni Foundation to develop lung cancer diagnostics for subtyping and predicting response to novel treatments using innovative methods to analyze cell-free DNA. The outcome of this work will have great potential to both guide cancer treatment decisions and to enable early lung cancer detection. We are grateful to the Kuni Foundation for their generous support of our pursuit to improve the lives of patients with lung cancer.

Welcome, Eden Cruikshank and Hunter Colegrove.
Eden joins the lab as our newest graduate student in the MCB program. She will be studying cancer genomes and developing new approaches to study cell-free DNA for her thesis research.
Hunter is a first year student in the Genome Sciences graduate program and joins us for his Winter 2022 Rotation. Hunter will be studying DNA repair pathway signatures in prostate cancer using circulating tumor DNA.

Congratulations to Anat Zimmer on receiving a Trainee Grant from the Brotman Baty Institute for Precision Medicine.
Anat’s project will involve the study of androgen receptor splice variants in prostate cancer using circulating tumor DNA. She will use computational and machine learning approaches to develop a signature for predicting ARv7 status.

Pushpa Itagi joins as a postdoctoral fellow, jointly mentored by Dr. Andrew Hsieh. She received her PhD in Computational Biology at the University of Kansas and was a visiting graduate researcher at the UCLA. She will work on bladder cancer genome and liquid biopsy projects, spearheading the collaboration between the Ha and Hsieh Labs, as well as the Bladder Cancer Group at Fred Hutch and UW.

Anat Zimmer is a new postdoctoral fellow in the lab. She received her PhD in computational biology at the Weizmann Institute before joining the ISB in Seattle. Now, in the Ha lab, she will work on cell-free DNA projects including development of new computational methods to study treatment response in patients with cancer.
Patty Galipeau joins the lab as a Research Project Manager overseeing the experimental wet lab operations and provide management support for the lab’s overall research program and collaborations. We are excited to have Patty bring her wealth of management, research experience, and leadership to help drive forward our mission.

Welcome to Abbey Thorpe and Adam Kreitzman.
Abbey is a graduate student in the Genome Sciences department at UW and is rotating in the lab this summer semester. She will be working on a project to analyze cell-free DNA ultra-low pass sequencing data in prostate cancer to identify transcriptional signatures in response to treatments.
Adam is an undergraduate student in his senior year at UCSD. He joins the lab for this summer and fall as an undergrad research assistant working on developing improved methods for cancer detection from ctDNA.

The Ha lab is awarded a CDMRP Department of Defense Prostate Cancer Research Program Idea Development Award to develop innovative approaches using cell-free DNA. This is a 3 year grant which we propose to develop new algorithms to study different layers of the cellular machinery that regulates genes using standard sequencing of ctDNA molecules. By employing machine learning techniques, we will combine different strategies of analyzing of cell-free data to classify the tumor subtypes and to identify tumor changes when treatments fail.

Congratulations to Adil for successfully completing his MSc degree in the UW Laboratory Medicine Graduate Program. Adil’s thesis was titled, "Accurate quantification of placental (fetal) fraction by tissue specific cell-free DNA analysis"

Congratulations to Robert Patton on receiving a Trainee Grant from the Brotman Baty Institute for Precision Medicine.
Robert’s project will involve the study of prostate cancer phenotype classification using circulating tumor DNA. He proposes use machine learning techniques to develop predictive model to analyze genomic and nucleosome features from cell-free DNA fragmentation patterns. The outcome of his work will have important applications in tumor monitoring and diagnostics for men with advanced prostate cancer.

A warm welcome to Eden Cruikshank and Mohamed Adil
Eden is a MCB graduate rotation student for this Winter 2021 semester. She will be developing models to detect breast cancer from DNA fragment analysis of circulationg tumor DNA.
Adil is a graduate student in the Master of Science in Laboratory Medicine program at UW Medicine. He will be completing his thesis research in the Ha Lab, under direct supervision with Dr. Jonathan Reichel and in close collaboration with Dr. Colin Pritchard and Shreeram Akilesh at UW Medicine.

A warm welcome to Robert Patton, who joins the lab as a postdoctoral fellow in the Computational Biology Program. He will be co-mentored by Dr. Peter Nelson of Fred Hutch.
Robert just finished his Ph.D. in theoretical biophysics at The Ohio State University. At Fred Hutch, he will leverage his unique expertise to study the cancer genomes of advanced prostate cancers from tissue and liquid biopsies.
Congratulations to Dr. Gavin Ha for being named a Top 40 Under 40 in Cancer in 2020, recognizing his achievements as a rising star and emerging leader in cancer research.

A warm welcome to Kelsey Luu and Hanna Liao
Kelsey is a Master’s student in the Harvard Biomedical Informatics and she is working in the lab as a Bioinformatics Intern to study the complex rearrangements of prostate cancer genomes.
Hanna is a MCB graduate rotation student for this Fall 2020 semester. She will be studying the transcriptional regulation of breast cancer and neuroblastoma from circulationg tumor DNA.
We are very grateful to the NCI for supporting Dr. Gavin Ha with a Transition Career Development Award (K22). This award will help support his research for the next 3 years to develop computational methods for emerging technologies to analyze tumor and cell-free DNA and will provide insights into the mechanisms underpinning treatment resistance in metastatic castration resistant prostate cancer. The K22 award will allow Gavin to further develop his scientific, professional, and leadership skills and prepare him to become a well-rounded independent investigator, leading a multi-disciplinary cancer research group.

We are very grateful for the support of The V Foundation for awarding our lab the V Scholar Grant to develop liquid biopies approaches for identifying molecular signatures of treatment resistance in advanced prostate cancer.
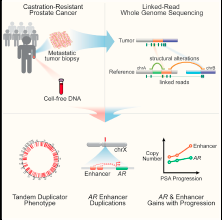
Here is the story and the description of the project.
This project aims to better characterize the genetic landscape of castration-resistant prostate cancer (CRPC) to uncover genomic alterations implicated in therapeutic resistance. We are using leading research approaches to leverage “liquid biopsies” to address the challenges of tissue inaccessibility and to study the complex evolving genomic landscape of CRPC during the course of therapy.
The ultimate goal of the proposed work is to help reduce mortality from advanced prostate cancer by using innovative approaches and exploiting non-invasive biomarkers in ctDNA to predict treatment outcomes. Metastatic castration-resistant prostate cancer (CRPC) is a lethal disease with no cure. Because the majority of metastases spread to the bone, obtaining tissue biopsies are painful. Furthermore, it is not always feasible to repeatedly perform surgical biopsies during the course of treatment to assess the cancer.
The proposed approaches aim to leverage liquid biopsies as a non-invasive and sensitive screen for circulating tumor DNA (ctDNA) in cancer from patient blood. The detection of ctDNA can be used for monitoring changes in disease burden such as during progression while on treatment or disease relapse. We also propose that ctDNA can be used to identify genetic signatures in CRPC, which can pinpoint specific vulnerabilities of the cancer to therapies.
The translational impact for successfully achieving the goals of this proposed research is three-fold: (1) the development of methods will help to accelerate the use ctDNA for clinical applications; (2) the identification of genomic signatures from ctDNA can be incorporated into the design of new biomarker-guided clinical trial for CRPC; (3) the insights into therapeutic response and resistance learned from this study may reveal new directions in diagnostics and treatment of CRPC. The ability to use minimally-invasive liquid biopsies has the potential to complement or replace tissue biopsies, which will transform the standard-of-care and improve the lives of men with prostate cancer.
This research requires the strong collaborative efforts between computational, experimental, and clinical scientists, embodying the paradigm of “computer-to-bench-to-bedside.” Our key collaborators and partners at the Fred Hutch, Seattle Cancer Care Alliance, and University of Washington Medicine will be instrumental to the success of this project.
A warm welcome to Minjeong Ko, a new Bioinformatics Analyst, and to Yuzhen Liu, an MCB graduate rotation student.
A warm welcome to rotation students Maggie Russell and Will Hannon.
Maggie and Will are graduate students in the MCB Program rotating for this Winter semester. They will be studying signatures of structural alterations in advanced prostate cancer.
A warm welcome to new lab member Anna Hoge.
Anna joins us as a Bioinformatics Analyst. She will be performing analysis on a variety of cancer datasets from on-going projects in the lab and through collaborations. We are delighted to have her start working on exciting research questions to study the genomes of advanced cancer patients.
A warm welcome to new lab member Jonathan Reichel.
Jonathan is a Lead Scientist at the Brotman Baty Institute (BBI) for Precision Medicine focused on cell-free DNA research. He is working with Dr. Colin Pritchard and the Ha Lab to develop new approaches for studying tissue-of-origin from circulating tumor DNA. He is facilitating its implementation into the BBI Clinical Diagnostic Platform.
A warm welcome to new lab member Eliza Barkan.
Eliza joins us as a graduate rotation student in the MCB Program for this spring semester. She will be studying signatures of structural alterations in advanced prostate cancer.
A warm welcome to the newest lab member Anna-Lisa Doebley.
Anna-Lisa Doebley is joining us as an MSTP student in the MCB Program. She will be developing and applying novel approaches to analyze cell-free DNA from advanced cancer patients.
A warm welcome to new lab members Katharine Chen and Megan Chiang.
Katharine joins us as a graduate rotation student in the MCB Program for this winter semester. She will be investigating approaches to analyze cell-free DNA from advanced cancer patients.
Megan join us as a bioinformatics intern from Northeastern University where she is completing her MSc. She will be analyzing tumor whole genome sequencing data from castration-resistant prostate cancer.
We are excited for them to start working on these exciting projects.
A computational biologist/bioinformatics analyst position in the laboratory of Dr. Gavin Ha and the Computational Biology Program is available immediately. We are seeking a highly motivated individual who is interested in studying the genetics and epigenetics of cancer using computational approaches. Candidates who are excited about large/complex ‘omics’ data analysis and cancer research are encouraged to apply. The position has a competitive salary with great benefits.
Please apply through the job posting here
Applicants must have at least a Bachelor’s degree in one of these disciplines:
- Computational biology, bioinformatics, computer science, data science, statistics, biostatistics, biomedical engineering, computer/electrical engineering, physics, or other related fields
Applicants should have some of the following skills and experience:
- Work well in team environments; strong communication skills; detail-oriented
- Strong programming experience (R, Python, Matlab, Java, C/C++, Perl or other languages for research)
- Experience with high performance computing environments or cloud computing environments is a plus
- Experience with analyzing sequencing data is considered an asset
- A background in cancer biology is considered an asset.
To apply, please submit your application with the following:
- Cover letter describing your experience and/or research interests
- CV
- Example of your code either as (1) an attachment or (2) link(s) to public repositories (e.g. GitHub)
A postdoctoral position in the laboratory of Dr. Gavin Ha and the Computational Biology Program is available immediately. We are seeking a highly motivated individual who is interested in studying cancer and understanding the genetic and epigenetic basis driving cancer progression. Candidates who are excited about large/complex ‘omics’ data analysis and methods development for cancer research are encouraged to apply. The position has a competitive salary and great benefits.
Please apply through the job posting here
Applicants must have a PhD in one of these disciplines:
- Computational biology, bioinformatics, computer science, data science, statistics, computer/electrical engineering, physics, or other related fields
Applicants should have some of the following skills and experience:
- Work well in team environments; strong communication/organization skills; detail-oriented
- Strong programming experience (R, Python, Matlab, Java, C/C++, Perl or other languages for research)
- Experience with high performance computing environments and cloud computing environments is a plus
- Experience with analyzing sequencing data is considered a strong asset
- Applicants must have a demonstrated publication track record.
- A background in cancer biology (esp in prostate or breast cancer) is considered a strong asset.
To apply, please submit your application with the following:
- A statement of research interests and past accomplishments
- CV
- Names and email addresses of three references
- Two representative publications or preprints (if available)
Check out the paper here
Under construction – Please check back later for updates.