Abstract
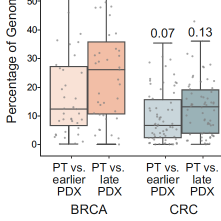
Genomic evolution of patient-derived xenografts (PDXs) may lead to their gradual divergence away of their tumors of origin. We previously reported the genomic evolution of the copy number (CN) landscapes of PDXs during their engraftment and passaging. However, whether PDX models are highly stable throughout passaging, or can evolve CNAs rapidly, remains controversial. Here, we reassess the genomic evolution of PDXs using DNA-based CN profiles. We find strong evidence for genomic evolution in the DNA-based PDX data: a median of ~10% of the genome is differentially altered between matched primary tumors (PTs) and PDXs across cohorts (range, 0% to 73% across all models). In 24% of the matched PT-PDX samples, over a quarter of the genome is differentially affected by CN alterations. Moreover, in matched analyses of PTs and their derived PDXs at multiple passages, later-passage PDXs are significantly less similar to their parental PTs than earlier-passage PDXs, indicative of genomic divergence. We conclude that PDX models indeed evolve throughout their derivation and propagation, and that the phenotypic consequences of this evolution ought to be assessed in order to determine its relevance to the proper application of these valuable cancer models.