Abstract
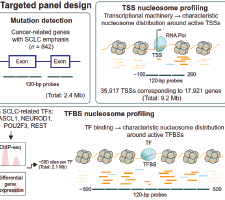
We report an approach for cancer phenotyping based on targeted sequencing of cell-free DNA (cfDNA) for small cell lung cancer (SCLC). In SCLC, differential activation of transcription factors (TFs), such as ASCL1, NEUROD1, POU2F3, and REST defines molecular subtypes. We designed a targeted capture panel that identifies chromatin organization signatures at 1535 TF binding sites and 13,240 gene transcription start sites and detects exonic mutations in 842 genes. Sequencing of cfDNA from SCLC patient-derived xenograft models captured TF activity and gene expression and revealed individual highly informative loci. Prediction models of ASCL1 and NEUROD1 activity using informative loci achieved areas under the receiver operating characteristic curve (AUCs) from 0.84 to 0.88 in patients with SCLC. As non-SCLC (NSCLC) often transforms to SCLC following targeted therapy, we applied our framework to distinguish NSCLC from SCLC and achieved an AUC of 0.99. Our approach shows promising utility for SCLC subtyping and transformation monitoring, with potential applicability to diverse tumor types.