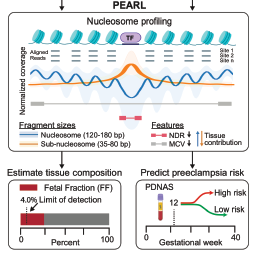
Preeclampsia is characterized by placental dysfunction and results in significant morbidity, but reliable early prediction remains challenging. We investigated whether clinically obtained prenatal cell-free DNA (cfDNA) screening (PDNAS) using whole-genome sequencing (WGS) data can be leveraged to predict preeclampsia risk early in pregnancy (≤16 weeks). Using 1,854 routinely collected clinical PDNAS samples (median, 12.1 weeks) with low-coverage (0.5×) WGS data, we developed a framework to quantify maternal and fetal tissue signatures using nucleosome accessibility, revealing early placental and endothelial dysfunction. These signatures informed a prediction model for preeclampsia risk, which achieved a validation performance of 0.85 area under the receiver operating characteristic curve (AUC) (81% sensitivity at 80% specificity) for preterm phenotypes several months prior to disease onset in a separate cohort of 831 consecutively collected samples, and subsequently confirmed in an external cohort of 141 samples (AUC 0.84, 79% sensitivity). We demonstrate that assessment of cfDNA nucleosome accessibility from early-pregnancy cfDNA sequence data enables the detection of early placental and endothelial-tissue aberrations and may aid in the determination of preeclampsia risk.
Preeclampsia risk prediction from prenatal cell-free DNA screening
Mohamed Adil, Teodora R Kolarova, Anna-Lisa Doebley, Leah A Chen, Cara L Tobey, Patricia Galipeau, Sam Rosen, Michael Yang, Brice Colbert, Robert D Patton, Thomas W Persse, Erin Kawelo, Jonathan B Reichel, Colin C Pritchard, Shreeram Akilesh, Christina M Lockwood, Gavin Ha+, Raj Shree+.
Nature Medicine
31(4),
1312-1318
(2025).